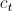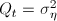Local level model
For example, suppose we have three observations $y_1,y_2,y_3$
Then
Measurement equation
State equation
where
To carry out MLE of the parameters we must form the log likelihood function. In time series analysis observations are typically not independent so to write the likelihood function, the standard approach is to use the prediction-error decomposition of the likelihood function. This done by factorizing the joint-density into a a product of conditional density functions.
For example, suppose we have three observations $y_1,y_2,y_3$
Then
$f(y_1,y_2,y_3) = f(y_3|y_2,y_1)f(y_2,y_1)$
$= f(y_3|y_2,y_1)f(y_2|y_1)f(y_1)$
In general for T observations we have
$L(y_1,y_2,...,y_T;\theta) = (\prod_{t=2}^{T}\ f(y_t|y_{t-1},..y_1))f(y_1)$
$= (\prod_{t=2}^{T}\ f(y_t|I_{t-1}))f(y_1)$
Where $I_t$ denotes all available information at date t.
Now recall from last time we had the following result
where implicit in the notation we have conditioned on all available information at date t-1.
which implies
Now define
Then the log likelihood function is given by
$L(\theta | y_1,..y_T; a_0, P_0) = $
$- \frac{nT}{2}ln(2\pi) - \frac{1}{2} \sum_{t=1}^{T} (ln|F_t(\theta)| + v_t(\theta)'F_t(\theta)^{-1}v_t(\theta))$
Which can then be maximised numerically with respect to the unknown parameters $\theta$.
Example
The R code below generates data from the local level model and estimates the variances by QMLE.
# local level model
# observation eq: yt = alphat + et
# state eq: alphat = alphat_1 + nt
# assume et is iidN(0,H)
# assume nt is iidN(0,Q)
# assume initial state alpha0 is N(a0,P0)
# initial conditions
a0 <- 0
P0 <- 10^6
# choose number of observations
n <- 250
# generate data
set.seed(1234)
H <- 10
Q <- 0.01
eta <-rnorm(n, mean = 0,sd = sqrt(Q))
alpha <- cumsum(eta)
epsilon <- rnorm(n, mean = 0, sd = sqrt(H))
y <- alpha + epsilon
#construct the negative loglikelihood function
neg.logl <- function(y,a0,P0,theta) {
#create variables
H <- exp(theta[1])
Q <- exp(theta[2])
#note we have reparameterized the variance to avoid the problem of negative estimates of the variances
#note we have reparameterized the variance to avoid the problem of negative estimates of the variances
n <- length(y)
a.tt_1 <- rep(0,n)
y.tt_1 <- rep(0,n)
v.tt <- rep(0,n)
a.tt <- rep(0,n)
P.tt_1 <- rep(0,n)
P.tt <- rep(0,n)
F.tt_1 <- rep(0,n)
Kt <- rep(0,n)
lht <- rep(0,n)
a.tt[1] <- a0
y.tt_1[1] <- a.tt[1]
P.tt_1[1] <- P0
# start filter
for(i in 2:n) {
# calculate initial guess of alphat based on all information from t_1
a.tt_1[i] <- a.tt[i-1]
# predicted yt given all information from t_1, and calculate the prediction error
y.tt_1[i] <- a.tt_1[i]
v.tt[i] <- y[i] - y.tt_1[i]
# calculate MSE of initial guess of alphat based on info from t_1
P.tt_1[i] <- P.tt[i-1] + Q
F.tt_1[i] <- P.tt_1[i] + H
# calculate the Kalman gain for date t
Kt[i] <- P.tt_1[i] * (F.tt_1[i])^(-1)
# combine prediction of yt with actual yt and initial guess of alpha t to obtain
# final estimate of alphat and the associated MSE for that estimate
a.tt[i] <- a.tt_1[i] + Kt[i] * v.tt[i]
P.tt[i] <- (1 - Kt[i]) * P.tt_1[i]
# calculate log of the density fn for observation i
lht[i] <- dnorm(v.tt[i], mean = 0, sd = sqrt(F.tt_1[i]), log = TRUE)
}
# calculate negative log likelihood fn
logl <- sum(lht)
return(-logl)
}
#estimate H and Q
#the optim() fn in R minimises the objective function which is why we calculated the negative log likelihood fn
# optim() requires starting values - which choose to use random start values
#the optim() fn in R minimises the objective function which is why we calculated the negative log likelihood fn
# optim() requires starting values - which choose to use random start values
start_val <- c(runif(1),runif(1))
output <- optim(start_val, neg.logl, method = "BFGS", y=y,a0=0,P0=10^6, hessian = T)
#get results
theta <- output$par
# remember we maximised w.r.t the log of the variances thus we need to take the exp() of theta
# remember we maximised w.r.t the log of the variances thus we need to take the exp() of theta
estim <- exp(theta)
names(estim) <- c("H.estim", "Q.estim")
# calculate the variance-covariance matrix
# note we don't need to take the minus here because we already did that when constructing the negative log likelihood
# note we don't need to take the minus here because we already did that when constructing the negative log likelihood
V <- solve(output$hessian)
You should get approximately 11.25 and 0.023 for the MLE of H and Q respectively, which are reasonably close to their true values.
You should get approximately 11.25 and 0.023 for the MLE of H and Q respectively, which are reasonably close to their true values.